Document Type : Review Article
Authors
- Nasim Almasian-Tehrani 1, 2
- Masoud Alebouyeh 2
- Shahnaz Armin 2
- Neda Soleimani 1
- Leila Azimi 2
- Roozbeh Shaker-Darabad 3
1 Department of microbiology and microbial biotechnology, faculty of life sciences and biotechnology, Shahid Beheshti University, Tehran, Iran.
2 Pediatric Infections Research Center, Research Institute for Children’s Health, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
3 Department of Biotechnology, Faculty of Advanced Sciences and Technologies, University of Isfahan, Isfahan, Iran
Abstract
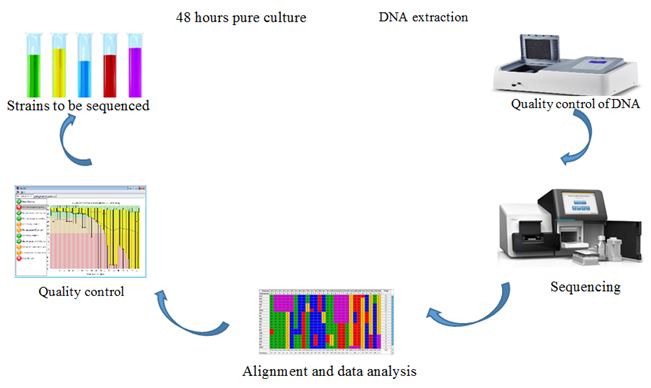
The main purpose of microbial typing is to evaluate the relationships between microbial isolates. Microbial typing can use for identifying the source of infection by detecting a clonal link between the strains. Moreover, it can analyze outbreaks, antimicrobial-resistant strains, and evaluate the effectiveness of control measures so, the efficiency of monitoring systems would increase. HAIs can affect hospitalized patients in all age ranges with any clinical situation, and lead to death. Molecular epidemiology is useful to determine genetic relatedness between isolated pathogens from patients, and design proper prevention plans to prevent infection through the hospital and community. Nowadays, typing methods for a wide range of bacterial strains are known as essential epidemiological tools to prevent and control infections in hospitals and communities. Although basic typing methods were more focused on phenotypic techniques like antibiogram and serotyping, new methods are based on molecular techniques including PCR-based methods and sequencing-based methods. Due to the high frequency of methods, choosing the right one for research applications seems difficult and requires basic knowledge about all of them. In this review, we aim to introduce the most useful and practical molecular typing techniques. Also, their utilization, advantages, and disadvantages were compared.
Graphical Abstract
Keywords
Main Subjects
Selected authors of this article by journal

Shahid Beheshti University of Medical Sciences |

Shahid Beheshti University of Medical Sciences |
| Google Scholar(H Index=18) | Google Scholar(H Index=15) |
Open Access
This article is licensed under a CC BY License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit: http://creativecommons.org/licenses/by/4.0/
Publisher’s Note
CMBR journal remains neutral with regard to jurisdictional claims in published maps and institutional afflictions.
Letters to Editor
Given that CMBR Journal's policy in accepting articles will be strict and will do its best to ensure that in addition to having the highest quality published articles, the published articles should have the least similarity (maximum 15%). Also, all the figures and tables in the article must be original and the copyright permission of images must be prepared by authors. However, some articles may have flaws and have passed the journal filter, which dear authors may find fault with. Therefore, the editor of the journal asks the authors, if they see an error in the published articles of the journal, to email the article information along with the documents to the journal office.
CMBR Journal welcomes letters to the editor ([email protected], [email protected]) for the post-publication discussions and corrections which allows debate post publication on its site, through the Letters to Editor. Critical letters can be sent to the journal editor as soon as the article is online. Following points are to be considering before sending the letters (comments) to the editor.
[1] Letters that include statements of statistics, facts, research, or theories should include appropriate references, although more than three are discouraged.
[2] Letters that are personal attacks on an author rather than thoughtful criticism of the author’s ideas will not be considered for publication.
[3] There is no limit to the number of words in a letter.
[4] Letter writers should include a statement at the beginning of the letter stating that it is being submitted either for publication or not.
[5] Anonymous letters will not be considered.
[6] Letter writers must include Name, Email Address, Affiliation, mobile phone number, and Comments.
[7] Letters will be answered as soon as possible.
- Ruppitsch W (2016) Molecular typing of bacteria for epidemiological surveillance and outbreak investigation/Molekulare Typisierung von Bakterien für die epidemiologische Überwachung und Ausbruchsabklärung. Die Bodenkultur: Journal of Land Management, Food and Environment 67(4):199-224. doi: https://doi.org/10.1515/boku-2016-0017
- Foxman B, Riley L (2001) Molecular epidemiology: focus on infection. American journal of epidemiology 153(12):1135-1141. doi:https://doi.org/10.1093/aje/153.12.1135
- Rodríguez-Morales AJ, Balbin-Ramon GJ, Rabaan AA, Sah R, Dhama K, Paniz-Mondolfi A, Pagliano P, Esposito S (2020) Genomic Epidemiology and its importance in the study of the COVID-19 pandemic. genomics 1:3
- Jensen AK, Björkman JT, Ethelberg S, Kiil K, Kemp M, Nielsen EM (2016) Molecular typing and epidemiology of human listeriosis cases, Denmark, 2002–2012. Emerging infectious diseases 22(4):625. doi:https://doi.org/10.3201/eid2204.150998
- Xiao L, Ryan UM (2007) Molecular epidemiology. In: Cryptosporidium and cryptosporidiosis. CRC press, pp 119-172
- Monegro AF, Muppidi V, Regunath H (2020) Hospital acquired infections. Statpearls [Internet])
- Alavi M, Rai M (2021) Antisense RNA, the modified CRISPR-Cas9, and metal/metal oxide nanoparticles to inactivate pathogenic bacteria. Cellular, Molecular and Biomedical Reports:52-59
- Azimi L, Alaghehbandan R, Asadian M, Alinejad F, Lari AR (2019) Multi-drug resistant Pseudomonas aeruginosa and Klebsiella pneumoniae circulation in a burn hospital, Tehran, Iran. GMS hygiene and infection control 14. doi: https://doi.org/10.3205/dgkh000317
- Soleimani N (2018) A Review of Designing New Vaccines to Prevent Hospital-Acquired Antibiotic-Resistant Infections. International Electronic Journal of Medicine 7(2):21-29. doi:https://doi.org/10.31661/iejm861
- Azimi L, Talebi M, Khodaei F, Najafi M, Lari AR (2016) Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis typing of carbapenemases producing Acinetobacter baumannii isolated from burn patients. Burns 42(2):441-445. doi:https://doi.org/10.1016/j.burns.2015.08.024
- Crabb HK, Allen JL, Devlin JM, Firestone SM, Wilks CR, Gilkerson JR (2018) Salmonella spp. transmission in a vertically integrated poultry operation: Clustering and diversity analysis using phenotyping (serotyping, phage typing) and genotyping (MLVA). PLoS One 13(7):e0201031. doi:https://doi.org/10.1371/journal.pone.0201031
- Ranjbar R, Karami A, Farshad S, Giammanco GM, Mammina C (2014) Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiologica 37(1):1-15
- Magalhães B, Valot B, Abdelbary MM, Prod'hom G, Greub G, Senn L, Blanc DS (2020) Combining standard molecular typing and whole genome sequencing to investigate Pseudomonas aeruginosa epidemiology in intensive care units. Frontiers in public health 8:3. doi:https://doi.org/10.1186/1471-2180-6-9
- Abbas-Al-Khafaji ZK, Aubais-aljelehawy Qh (2021) Evaluation of antibiotic resistance and prevalence of multi-antibiotic resistant genes among Acinetobacter baumannii strains isolated from patients admitted to al-yarmouk hospital. Cellular, Molecular and Biomedical Reports:60-68
- Christensen H, Bossé J, Angen Ø, Nørskov-Lauritsen N, Bisgaard M (2020) Immunological and molecular techniques used for determination of serotypes in Pasteurellaceae. Methods in Microbiology 47:117-149. doi:https://doi.org/10.1016/bs.mim.2020.01.002
- Armin S, Fallah F, Karimi A, Rashidan M, Shirdust M, Azimi L (2017) Genotyping, antimicrobial resistance and virulence factor gene profiles of vancomycin resistance Enterococcus faecalis isolated from blood culture. Microbial pathogenesis 109:300-304. doi:https://doi.org/10.1016/j.micpath.2017.05.039
- Kaufmann M, Pitcher D, Pitt T (2018) Ribotyping of bacterial genomes. In: Methods in Practical Laboratory Bacteriology. CRC Press, pp 123-138
- Van Belkum A, Welker M, Pincus D, Charrier J-P, Girard V (2017) Matrix-assisted laser desorption ionization time-of-flight mass spectrometry in clinical microbiology: what are the current issues? Annals of laboratory medicine 37(6):475-483. doi:https://doi.org/10.3343/alm.2017.37.6.475
- Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E (2018) Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clinical microbiology and infection 24(4):335-341. doi:https://doi.org/10.1016/j.cmi.2017.10.013
- Lagha R, Abdallah FB, ALKhammash AA, Amor N, Hassan MM, Mabrouk I, Alhomrani M, Gaber A (2021) Molecular characterization of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from King Abdulaziz Specialist Hospital at Taif City, Saudi Arabia. Journal of Infection and Public Health 14(1):143-151. doi:https://doi.org/10.1016/j.jiph.2020.12.001
- Singh A, Goering RV, Simjee S, Foley SL, Zervos MJ (2006) Application of molecular techniques to the study of hospital infection. Clinical microbiology reviews 19(3):512-530. doi:https://doi.org/10.1128/CMR.00025-05
- Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, Kato H, Kai K, Arakawa Y (2003) PCR typing of genetic determinants for metallo-β-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. Journal of Clinical Microbiology 41(12):5407-5413. doi:https://doi.org/10.1128/JCM.41.12.5407-5413
- Van Belkum A (1994) DNA fingerprinting of medically important microorganisms by use of PCR. Clinical Microbiology Reviews 7(2):174-184. doi:https://doi.org/10.1128/CMR.7.2.174
- Van Belkum A, Tassios P, Dijkshoorn L, Haeggman S, Cookson B, Fry N, Fussing V, Green J, Feil E, Gerner‐Smidt P (2007) Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clinical Microbiology and Infection 13:1-46. doi:https://doi.org/10.1111/j.1469-0691.2007.01786.x
- Asadian M, Azimi L, Alinejad F, Ostadi Y, Lari AR (2019) Molecular characterization of Acinetobacter baumannii isolated from ventilator-associated pneumonia and burn wound colonization by random amplified polymorphic DNA polymerase chain reaction and the relationship between antibiotic susceptibility and biofilm production. Advanced biomedical research 8. doi:https://doi.org/10.4103/abr.abr_256_18
- Bilung LM, Pui CF, Su’ut L, Apun K (2018) Evaluation of BOX-PCR and ERIC-PCR as molecular typing tools for pathogenic Leptospira. Disease markers 2018. doi:https://doi.org/10.1155/2018/1351634
- Le Flèche P, Jacques I, Grayon M, Al Dahouk S, Bouchon P, Denoeud F, Nöckler K, Neubauer H, Guilloteau LA, Vergnaud G (2006) Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC microbiology 6(1):1-14. doi:https://doi.org/10.1186/1471-2180-6-9
- Ardakani MA, Ranjbar R (2016) Molecular typing of uropathogenic E. coli strains by the ERIC-PCR method. Electronic physician 8(4):2291. doi:https://doi.org/10.19082/2291
- Schmitt S, Stephan R, Huebschke E, Schaefle D, Merz A, Johler S (2020) DNA microarray-based characterization and antimicrobial resistance phenotypes of clinical MRSA strains from animal hosts. Journal of Veterinary Science 21(4). doi:https://doi.org/10.4142/jvs.2020.21.e54
- Gaiarsa S, Batisti Biffignandi G, Esposito EP, Castelli M, Jolley KA, Brisse S, Sassera D, Zarrilli R (2019) Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) schemes. Frontiers in microbiology 10:930. doi:https://doi.org/10.3389/fmicb.2019.00930
- Scholz CF, Jensen A (2017) Development of a Single Locus Sequence Typing (SLST) scheme for typing bacterial species directly from complex communities. In: Bacterial Pathogenesis. Springer, pp 97-107. doi:https://doi.org/10.1007/978-1-4939-6673-8_7
- Kwok P-Y (2001) Methods for genotyping single nucleotide polymorphisms. Annual review of genomics and human genetics 2(1):235-258. doi:https://doi.org/10.1146/annurev.genom.2.1.235
- Hallin M, Deplano A, Struelens MJ (2012) Molecular typing of bacterial pathogens: a tool for the epidemiological study and control of infectious diseases. In: New frontiers of molecular epidemiology of infectious diseases. Springer, pp 9-25. doi:https://doi.org/10.1007/978-94-007-2114-2_2
- Tümmler B (2020) Molecular epidemiology in current times. Environmental Microbiology 22(12):4909-4918. doi:https://doi.org/10.1111/1462-2920.15238
- Prado-Vivar MB, Ortiz L, Reyes J, Villacis E, Fornasini M, Baldeon ME, Cardenas PA (2019) Molecular typing of a large nosocomial outbreak of KPC-producing bacteria in the biggest tertiary-care hospital of Quito, Ecuador. Journal of global antimicrobial resistance 19:328-332. doi:https://doi.org/10.1016/j.jgar.2019.05.014
- Sharma-Kuinkel BK, Rude TH, Fowler VG (2014) Pulse field gel electrophoresis. In: The Genetic Manipulation of Staphylococci. Springer, pp 117-130
- García H, Cervantes-Luna B, González-Cabello H, Miranda-Novales G (2018) Risk factors for nosocomial infections after cardiac surgery in newborns with congenital heart disease. Pediatrics & Neonatology 59(4):404-409. doi:https://doi.org/10.1186/s12879-020-4769-6
- Corcione S, Pensa A, Castiglione A, Lupia T, Bortolaso B, Romeo MR, Stella M, Rosa FGD (2021) Epidemiology, prevalence and risk factors for infections in burn patients: Results from a regional burn centre’s analysis. Journal of Chemotherapy 33(1):62-66. doi:https://doi.org/10.1080/1120009X.2020.1780776
- Sikora A, Zahra F (2021) Nosocomial infections. StatPearls [Internet])
- Baran I, Aksu N (2016) Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann Clin Microbiol Antimicrob 15:20. doi:https://doi.org/10.1186/s12941-016-0136-2